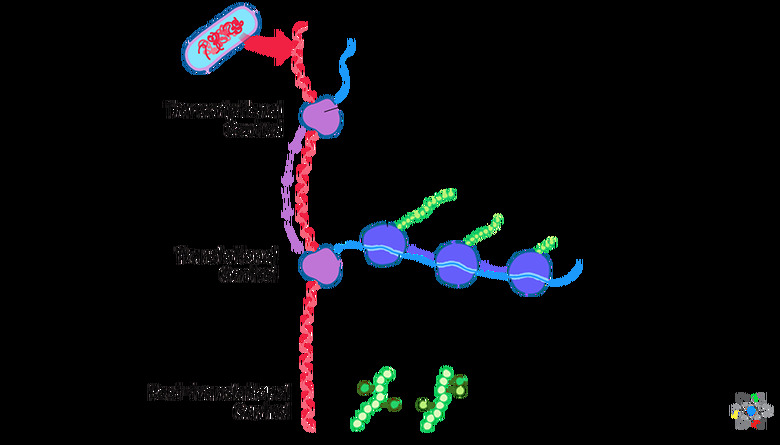
Gene Expression In Prokaryotes
Prokaryotes are small, single-celled living organisms. They are one of two common cell types: prokaryotic and **eukaryotic**.
Since prokaryotic cells do not have a nucleus or organelles, gene expression happens out in the open cytoplasm and all the stages can happen simultaneously. Although prokaryotes are simpler than eukaryotes, controlling gene expression is still crucial for their cellular behavior.
Genetic Information in Prokaryotes
Genetic Information in Prokaryotes
The two domains of prokaryotes are Bacteria and Archaea. Both lack a defined nucleus, but they still have a genetic code and nucleic acids. Although there are no complex chromosomes like those that you would see in eukaryotic cells, prokaryotes have circular pieces of deoxyribonucleic acid (DNA) located in the nucleoid.
However, there is no membrane around the genetic material. In general, prokaryotes have fewer non-coding sequences in their DNA compared to eukaryotes. This may be due to prokaryotic cells being smaller and having less space for a DNA molecule.
The nucleoid is simply the region where the DNA lives in the prokaryotic cell. It has an irregular shape and can vary in size. In addition, the nucleoid is attached to the cell membrane.
Prokaryotes can also have circular DNA called _plasmids_. It is possible for them to have one or more plasmids in a cell. During cell division, prokaryotes can go through DNA synthesis and the separation of plasmids.
Compared to the chromosomes in eukaryotes, plasmids tend to be smaller and have less DNA. In addition, plasmids can replicate on their own without other cellular DNA. Some plasmids carry the codes for nonessential genes, such as those that give bacteria their antibiotic resistance.
In certain cases, plasmids are also able to move from one cell to another cell and share information like antibiotic resistance.
Stages in Gene Expression
Stages in Gene Expression
Gene expression is the process through which the cell translates the genetic code into amino acids for protein production. Unlike in eukaryotes, the two main stages, which are transcription and translation, can happen at the same time in prokaryotes.
During transcription, the cell translates DNA into a messenger RNA (mRNA) molecule. During translation, the cell makes the amino acids from the mRNA. The amino acids will make up the proteins.
Both transcription and translation happen in the prokaryote's cytoplasm. By having both processes happen at the same time, the cell can make a large amount of protein from the same DNA template. If the cell does not need the protein any longer, then transcription can stop.
Transcription in Bacterial Cells
Transcription in Bacterial Cells
The goal of transcription is to create a complementary ribonucleic acid (RNA) strand from a DNA template. The process has three parts: initiation, chain elongation and termination.
In order for the initiation phase to occur, the DNA has to unwind first and the area where this happens is the transcription bubble.
In bacteria, you will find the same RNA polymerase responsible for all transcription. This enzyme has four subunits. Unlike eukaryotes, prokaryotes do not have transcription factors.
Transcription: Initiation Phase
Transcription: Initiation Phase
Transcription starts when the DNA unwinds and RNA polymerase binds to a promoter. A promoter is a special DNA sequence that exists at the beginning of a specific gene.
In bacteria, the promoter has two sequences: -10 and -35 elements. The -10 element is where the DNA usually unwinds, and it is located 10 nucleotides from the initiation site. The -35 element is 35 nucleotides from the site.
RNA polymerase relies on one DNA strand to be the template as it builds a new strand of RNA called the RNA transcript. The resulting RNA strand or primary transcript is almost the same as the non-template or coding DNA strand. The only difference is that all of the thymine (T) bases are uracil (U) bases in RNA.
Transcription: Elongation Phase
Transcription: Elongation Phase
During the chain elongation phase of transcription, RNA polymerase moves along the DNA template strand and makes an mRNA molecule. The RNA strand gets longer as more nucleotides are added.
Essentially, RNA polymerase walks along the DNA stand in the 3' to 5' direction to accomplish this. It is important to note that bacteria can create polycistronic mRNAs that code for multiple proteins.
Transcription: Termination Phase
Transcription: Termination Phase
During the termination phase of transcription, the process stops. There are two types of termination phases in prokaryotes: Rho-dependent termination and Rho-independent termination.
In Rho-dependent termination, a special protein factor called Rho interrupts transcription and terminates it. The Rho protein factor attaches to the RNA strand at a specific binding site. Then, it moves along the strand to reach the RNA polymerase in the transcription bubble.
Next, Rho pulls apart the new RNA strand and DNA template, so transcription ends. RNA polymerase stops moving because it reaches a coding sequence that is the transcription stop point.
In Rho-independent termination, the RNA molecule makes a loop and detaches. The RNA polymerase reaches a DNA sequence on the template strand that is the terminator and has many cytosine (C) and guanine (G) nucleotides. The new RNA strand starts to fold up into a hairpin shape. Its C and G nucleotides bind. This process stops the RNA polymerase from moving.
Translation in Bacterial Cells
Translation in Bacterial Cells
Translation creates a protein molecule or polypeptide based on the RNA template created during transcription. In bacteria, translation can happen right away, and sometimes it starts during transcription. This is possible because prokaryotes do not have any nuclear membranes or any organelles to separate the processes.
In eukaryotes, things are different because transcription occurs in the nucleus, and translation is in the cytosol, or intracellular fluid, of the cell. A eukaryote also uses mature mRNA, which is processed before translation.
Another reason why translation and transcription can happen at the same time in bacteria is that the RNA does not need the special processing seen in eukaryotes. The bacterial RNA is ready for translation immediately.
The mRNA strand has groups of nucleotides called codons. Each codon has three nucleotides and codes for a specific amino acid sequence. Although there are only 20 amino acids, cells have 61 codons for amino acids and three stop codons. AUG is the start codon and begins translation. It also codes for the amino acid methionine.
Translation: Initiation
Translation: Initiation
During translation, the mRNA strand acts as a template for making amino acids that become proteins. The cell decodes the mRNA to accomplish this.
Initiation requires transfer RNA (tRNA), a ribosome and mRNA. Each tRNA molecule has an anticodon for an amino acid. The anticodon is complementary to the codon. In bacteria, the process starts when a small ribosomal unit attaches to the mRNA at a Shine-Dalgarno sequence.
The Shine-Dalgarno sequence is a special ribosomal binding area in both bacteria and archaea. You usually see it about eight nucleotides from the start codon AUG.
Since bacterial genes can have transcription happen in groups, one mRNA may code for many genes. The Shine-Dalgarno sequence makes it easier to find the start codon.
Translation: Elongation
Translation: Elongation
During elongation, the chain of amino acids becomes longer. The tRNAs add amino acids to make the polypeptide chain. A tRNA starts working in the P site, which is a middle part of the ribosome.
Next to the P site is the A site. A tRNA that matches the codon can go to the A site. Then, a peptide bond can form between the amino acids. The ribosome moves along the mRNA, and the amino acids form a chain.
Translation: Termination
Translation: Termination
Termination happens because of a stop codon. When a stop codon enters the A site, the process of translation stops because the stop codon does not have a complementary tRNA. Proteins called release factors that fit into the P site can recognize the stop codons and prevent peptide bonds from forming.
This happens because the release factors can make enzymes add a water molecule, which makes the chain separate from tRNA.
Translation and Antibiotics
Translation and Antibiotics
When you take some antibiotics to treat an infection, they may work by disrupting the translation process in bacteria. The goal of antibiotics is to kill the bacteria and stop them from reproducing.
One way they accomplish this is to affect the ribosomes in bacterial cells. The drugs can interfere with mRNA translation or block the ability of the cell to make peptide bonds. Antibiotics can bind to the ribosomes.
For instance, one type of antibiotic called tetracycline can enter the bacterial cell by crossing the plasma membrane and building up inside the cytoplasm. Then, the antibiotic can bind to a ribosome and block translation.
Another antibiotic called ciprofloxacin affects the bacterial cell by targeting an enzyme responsible for unwinding the DNA to allow replication. In both cases, human cells are spared, which allows people to use antibiotics without killing their own cells.
Related topic: _multicellular organisms_
Post-Translation Protein Processing
Post-Translation Protein Processing
After translation is over, some cells continue processing the proteins. Post-translational modifications (PTMs) of proteins allow bacteria to adapt to their environment and control cellular behavior.
In general, PTMs are less common in prokaryotes than eukaryotes, but some organisms have them. Bacteria can modify proteins and reverse the processes, too. This gives them more versatility and allows them to use protein modification for regulation.
Protein Phosphorylation
Protein Phosphorylation
_Protein phosphorylation_ is a common modification in bacteria. This process involves adding a phosphate group to the protein, which has phosphorus and oxygen atoms. Phosphorylation is essential for protein function.
However, phosphorylation can be temporary because it is reversible. Some bacteria can use phosphorylation as part of the process to infect other organisms.
Phosphorylation that occurs on the serine, threonine and tyrosine amino acid side chains is called Ser/Thr/Tyr phosphorylation.
Protein Acetylation and Glycosylation
Protein Acetylation and Glycosylation
In addition to phosphorylated proteins, bacteria can have acetylated and glycosylated proteins. They may also have methylation, carboxylation and other modifications. These modifications play an important role in cell signaling, regulation and other processes in bacteria.
For instance, Ser/Thr/Tyr phosphorylation helps bacteria respond to changes in their environment and increase the chances of survival.
Research shows that metabolic changes in the cell are associated with Ser/Thr/Tyr phosphorylation, which indicates that bacteria can respond to their environment by changing their cellular processes. Moreover, post-translational modifications help them react quickly and efficiently. The ability to reverse any changes also provides significant control.
Gene Expression in Archaea
Gene Expression in Archaea
Archaea use gene expression mechanisms that are more similar to eukaryotes. Although archaea are prokaryotes, they have some things in common with eukaryotes, such as gene expression and gene regulation. The processes of transcription and translation in archaea also have some similarities with bacteria.
For instance, both archaea and bacteria have methionine as the first amino acid and AUG as the start codon. On the other hand, both archaea and eukaryotes have a TATA box, which is a DNA sequence in the promoter area that shows where to decode the DNA.
Translation in archaea resembles the process seen in bacteria. Both types of organisms have ribosomes that consist of two units: the 30S and 50S subunits. In addition, they both have polycistronic mRNAs and and Shine-Dalgarno sequences.
There are multiple similarities and differences among bacteria, archaea and eukaryotes. However, they all rely on gene expression and gene regulation to survive.
References
- Lumen: Prokaryotic and Eukaryotic Gene Regulation
- Lumen: Regulation of Gene Expression
- Biology LibreTexts: Prokaryotic Versus Eukaryotic Gene Expression
- OpenTextBC: Prokaryotic Gene Regulation
- Lumen: Prokaryotic Transcription
- Pearson: Transcription and Translation in Cells
- Lumen: Prokaryotic Transcription and Translation
- Carleton College: Translation: Messenger RNA Translated Into Protein
- Frontiers in Microbiology: Regulatory Potential of Post-Translational Modifications in Bacteria
- Frontiers in Microbiology: Phosphoproteome of the Cyanobacterium Synechocystis Sp. Pcc 6803 and Its Dynamics During Nitrogen Starvation
Cite This Article
MLA
Bandoim, Lana. "Gene Expression In Prokaryotes" sciencing.com, https://www.sciencing.com/gene-expression-in-prokaryotes-13717692/. 21 May 2019.
APA
Bandoim, Lana. (2019, May 21). Gene Expression In Prokaryotes. sciencing.com. Retrieved from https://www.sciencing.com/gene-expression-in-prokaryotes-13717692/
Chicago
Bandoim, Lana. Gene Expression In Prokaryotes last modified August 30, 2022. https://www.sciencing.com/gene-expression-in-prokaryotes-13717692/